A bioinformatics pipeline for iCLIP data analysis
A complete pipeline from sequencing reads to RBP binding sites
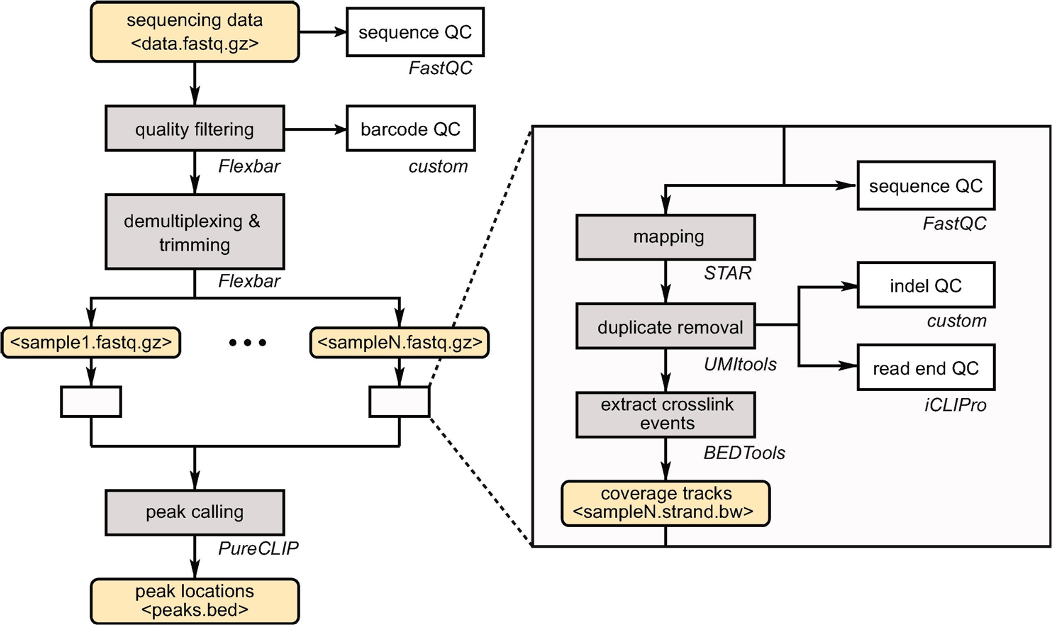
Precise knowledge on the binding sites of an RNA-binding protein (RBP) is key to understand (post-) transcriptional gene regulation. This information can be obtained from individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) experiments. In this project we discribed the complete bioinformatic data analysis workflow to reliably detect RBP binding sites from iCLIP data. The workflow covers all steps from the initial quality control of the sequencing reads up to peak calling and quantification of RBP binding. For each tool, we explain the specific requirements for iCLIP data analysis and suggest optimized parameter settings.
The full manuscript can be found at Methods and an updated version of the binding site definition part is published as the R/ Bioconductor packge BindingSiteFinder Bioconductor page.